Correcting and adjusting NAD pathways
So about the pathways I uploaded on Wikipathways last week. There were some small corrections still to be made about them. There were 2 main issues: The metabolites that go into or out of a reaction with the main metabolites have to be grouped together and the data nodes have to be aligned correctly.
So this is how to tackle both of the problems. First we need to select the non-grouped metabolites that belong to each other: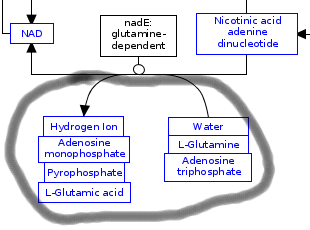
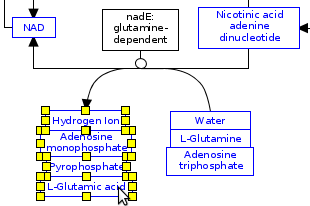
Here I push “Ctrl+g” to group the selected items together. Now that they are grouped you need to move the arrow from where it is bound to one metabolite to the box around the group:
Now that this is done we can start aligning the individual metabolites. this can be done by selecting all the metabolites inside the box and going to the top menu bar of pathvisio or Wikipathways (right side).
Using these 6 aligning button, I made my data nodes more readable (I mainly used the 1st and 3de one). Keeping in mind that it will adjust the data nodes to the largest data node:
Now we have a nice group of metabolites that belong or come from the same reaction and align correctly with each other so it’s more readable for everyone.
Using python rdflib
I am currently trying to understand the different formats for rdf, one way to do this is by making a rdf myself. Since I am more practiced with python, i’ll be using the rdflib a python module to build my own rdf about Saxophone (the instrument I used to play).
So first of all I am trying to get the information from dbpedia and store it in a specific format (either n-triples or turtle). After a few tutorials:
http://semanticweb.org/wiki/Getting_data_from_the_Semantic_Web; http://www.michelepasin.org/blog/2011/07/18/inspecting-an-ontology-with-rdflib/.
and some trial and error I finally managed to download the dbpedia about Saxophones and converted it into the specific format required. I know you can get any rdf format from dbpedia directly.
Here you have to make sure that the dbpedia link say “resource” not “page” which is the redirected link of the “resource” link. Also the format can be changed in all the required formats:
N-triples = nt
Turtle = turtle
Notation 3 = n3
RDf xml = rdf+xml
N-quads = nq
So now I know how to convert one RDF format to another. So now I need to make my own, by inserting my own subject, predicate and objects.
This can be achieved by using the “graph.add((sub,pre,obj))”. Again here is a useful manual about rdflib: http://rdflib.readthedocs.org/en/3.2.0/gettingstarted.html. Though honestly its not that clear in the beginning. In the next example i made 2 lines of triples and added in the nt format:
Again here I can change the format to any of the format that were mentioned above. below is an example of how the “nt” format looks like:
<http://rdflib.net/test/Saxophone> <http://rdflib.net/test/invented_by> "mr. Sax".
<http://rdflib.net/test/Saxophone> <http://rdflib.net/test/is_a> "instrument".
Now all I have to do is making a command based interactive script that allows me to enter the information I want manually and add it to the rdf file.
Since This was a test i didnt pay much attention on the predicates. Put I am currently trying to understand which predicates suppose to be used when. A few website I am currently studying:
http://xmlns.com/foaf/spec/
http://www.heppnetz.de/grprofile/
http://www.w3.org/TR/vcard-rdf/
And other related subject like: rdf, rdfs, dc, dcterms etc.
Geen opmerkingen:
Een reactie posten