Hi everyone,
I am Cizar Almalak and a few weeks back I started my internship here in Maastricht University. I choose to do my internship here in Maastricht because I was quite interested In the Semantic web, which is big issue here. Through Chris Evelo i came in contact with Andra Waagmeester, who is currently my supervisor.
The goal of my project exist of multiple steps. First I have to understand how Wikipathways works and the concept of Semantic Web. Secondly is to write a script that can convert RDF into GPML and thirdly is to write a script that can convert one RDF format into another RDF format.
So to understand how Wikipathways works, I have to practice by making some pathways myself, starting with the pathway tutorial on Wikipathways: http://wikipathways.org/index.php/Help:Tutorial.
After doing this tutorial, It was time to take a real pathway from different database than Wikipathways and see whether it was possible for me to reproduce it for Wikipathways. Since this will be my first time making a pathway for Wikipathways, instead of putting the pathway directly on on Wikipathways, I made it with pathvisio. Which has the same interface as Wikipathways and can export the data in GPML, which Wikipathways can read.
So in the following example i’ll be showing how I did the NAD biosynthesis pathway in E.coli. I got the pathway from Biocyc (source: http://biocyc.org/ECOLI/NEW-IMAGE?type=PATHWAY&object=PYRIDNUCSYN-PWY).
At fist I got the latest version of pathvisio (3).
So the first step when you have pathvisio, you need to download the required data from Wiki pathways, about the organism you are using (Genes and metabolite database). I ashieved this by going to the following link: http://www.pathvisio.org/downloads/download-bridgedbs/ or just going to www.pathvisio.org >> upper menu bar >> downloads >> download mapping databases:
And then I select the species that would like to use (E.coli for this example). For metabolites there is only a human one, which you can use on every specie.
After I downloaded the required databases I needed to upload them in pathvisio. This can be done by going to the upper panel of pathvisio and choosing: Data >> Select gene database and Data >> Select metabolite database.
So now that pathvisio is ready, I can start with the pathway. Before I started the pathway I went to the box that says “title” on the upper left corner and double clicked it. a menu will pop-up.
On the pop-up you will see on the top 3 options: Comments, Literature and Properties. In properties I put the title and organism I’m working on.
In the comments menu I put the source from where I tool the pathway.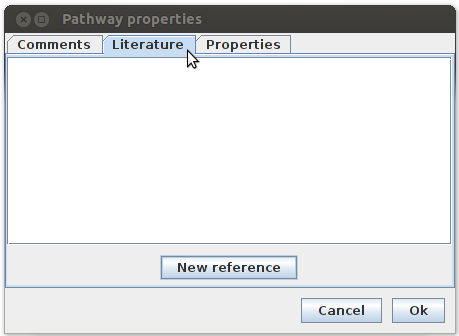
In the reference menu I put all the references that were associated with this pathway. I achieved this by pushing the new reference button below, which will open another pop-up menu:
By entering the pubmed id of the article and pushing the button on the right (Query PubMed), all the required information from the publication will be uploaded correctly.
Now that i have done this, I started building the pathway, based on the BioCyc NAD biosynthesis pathway of E.coli.
I started first creating all proteins/enzymes/metabolites data nodes without the interactions. For proteins and enzymes I used the gene data nodes and for the metabolites the metabolite data node. These data nodes can be found under “Objects” on the right side panel of pathvisio:
Just creating the data nodes is not enough, each protein/enzyme/metabolite has to be annotated. This is the tricky part since, some of the them have synonyms and in Wikipathway as pathvisio only recognise the primary name. So extra research is required to find the primary name. So first I double click on one of the data nodes and a data node property list will pop-up:
In this pop-up menu I entered in the search field the name of the protein/enzyme/metabolite in question and pushed search. If there is no hit, it means that this protein/enzyme/metabolite is not annotated or it is not its primary name. But in this case this is the primary name of the metabolite and I got another pop-up minute that shows me all the hits that were found:
So here I choose the metabolite in question, all other hits are similar but not the metabolite i was searching for. After I made the selection I pressed “ok” and the my metabolite in question was annotated based on the database I selected: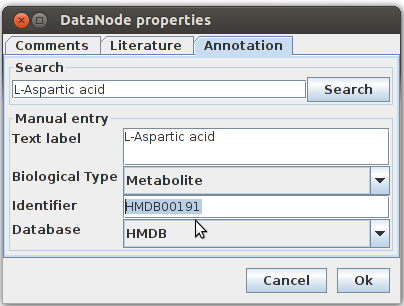
Any extra information or reference about the annotated protein/enzyme/metabolite can be added in the Comments or Literature tab on top of the data node property pop-up.
So when I had all the data nodes annotated, it was time to add the interactions between each data node. Again like the data nodes the interactions can be found on the right side panel under the data nodes section “Basic interactions” & “MIM interactions”:
I used “Basic interactions” when the type of interaction is not clear or not located in the “MIM interaction” list and “MIM interactions” when it is clearly stated what kind of interaction is involved between the 2 products. There are a few problems that occurred to me when trying to build this pathway. One is that when you have an interacting product with the main product, you have to use 2 different lines example:
The circles are 2 separate lines binding to a single place, to illustrate the involvement of the metabolite oxygen with L-Aspartic acid and the rest product hydrogen peroxide and H+. Secondly is that sometimes the program bugs and all data nodes and interactions scatter. This happens mainly when you try to move all of them at once.
Now that i have done all the interactions I am basically done, all I have to do now is put them in a nice order to make it more readable.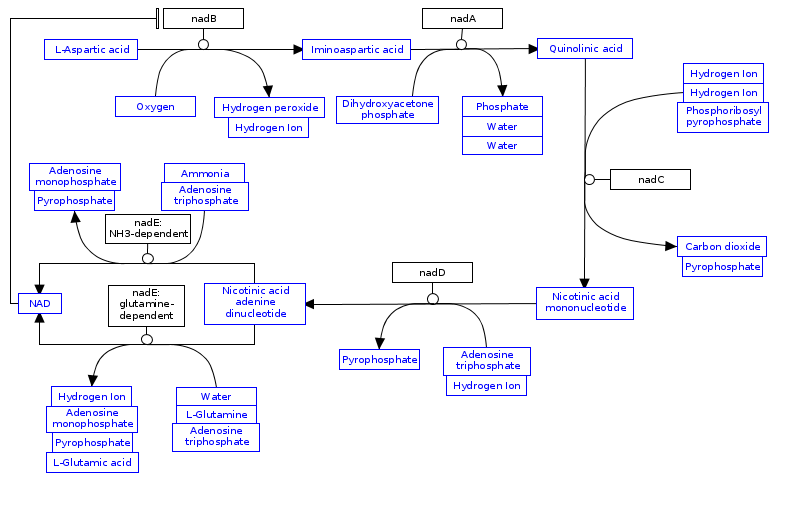
Using the top menu bar from pathvisio and selecting File >> Export: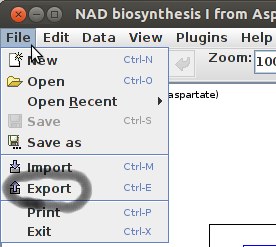
A pop-up menu will appear. using the new menu, you can give name to the file in the “File Name” field, and choose a format in the “Files of Type:” field. Currently i required the gpml format, to upload on Wikipathways.
After exporting the file into a gpml format I can now upload it in Wikipathways. So first i went to the Wikipathway website (http://wikipathways.org/index.php/WikiPathways) and choose create from the left side panel (circled). 
In the new window I can choose to create a new path or upload a new pathway in a certain format. 
After I uploaded the pathway, Everyone can see the pathways on Wikipathways and make adjustments to them. Also when the pathway is uploaded, I added a description to the pathway, so that people know what the pathway is about.
The description can be added under the header description under the pathway: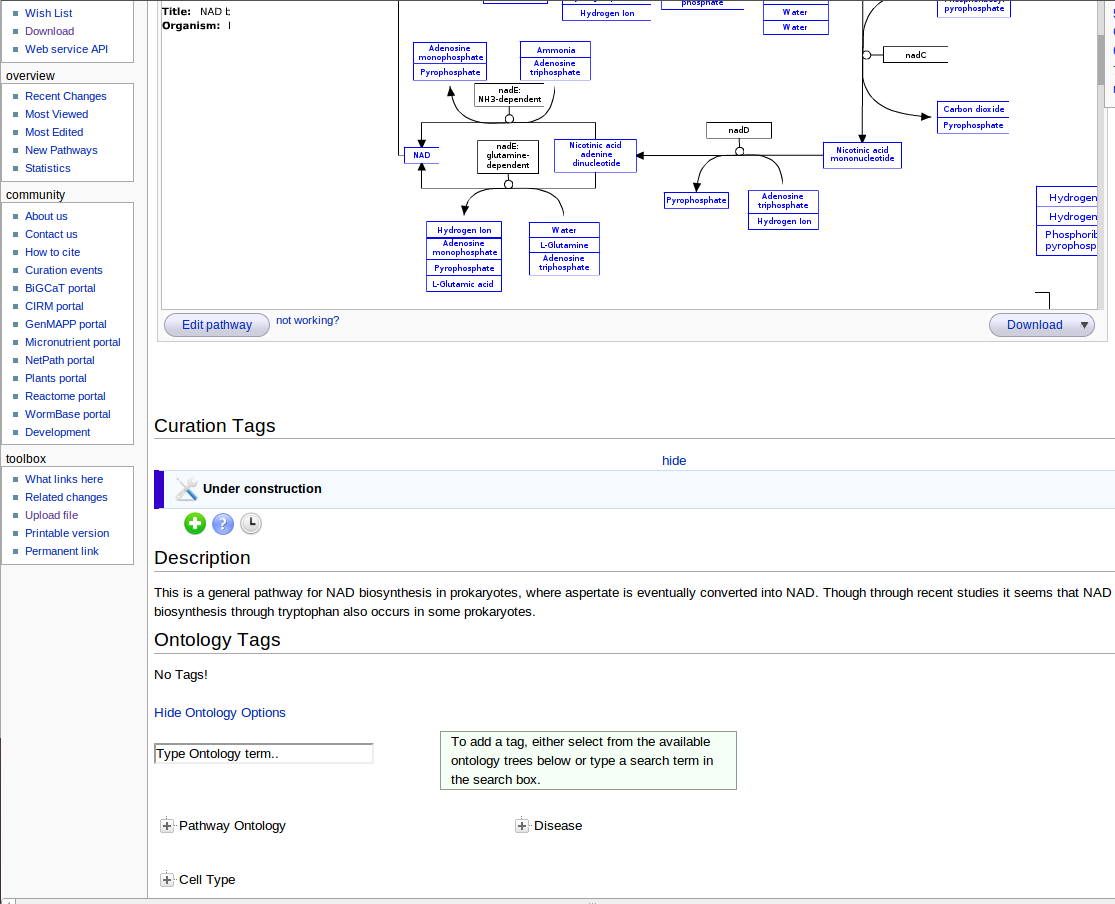
Also more information can be added to the pathway under different headers, if the information is available.
Geen opmerkingen:
Een reactie posten