Wikipathways is an open, public platform dedicated to the curation of biological pathways.
Some of the annotations were GeneOntology IDs. This is not prefered in Wikipathways (EC numbers as well). If the search function when trying to annotate a certain gene, does not find the gene it could be because of a few things:
- The gene is not located in the databases incorporated in Wikipathways.
- The gene name that is used is not the primary name of the gene (only primary names can be searched).
- The name was not typed correctly.
- Or the gene does not exist, which in this case it can not be annotated.
In the case of point 1, 2 or 3 extra research is required. I will show a small example about how I tackled these points.
Fist we go to the the Wikipathway webpage: http://wikipathways.org/index.php/WikiPathways.
From there I entered the name of my pathway in the search engine.

After we entered the name and pushed "Search" we can add the species to narrow down the search.

Now I choose the to be adjusted NAD pathway (NAD salvage pathway I). After I have selected pathway you can edit them by pushing the "Edit pathway" button on the lower left of the pathway screen. Now I double click on the data node that I want to adjust.
In the mean time I go to the original source from where I took the pathway. In this case it was: http://biocyc.org/ECOLI/NEW-IMAGE?type=PATHWAY&object=PYRIDNUCSAL-PWY&detail-level=2. Here I can see what the the name of the gene/enzyme/protein was before I annotated it. In this case it was also "NAD+ diphosphatase". Keep in mind the reason I am re-annotating this enzyme, is because GO is not preferred in Wikipathways because more than 1 gene can be linked to one GOterm.
So the first step is to try and find this enzyme/gene product a different database like ensembl (ensembl bacteria), uniprot etc. If like in this case we do not find a hit, we try to look whether it is a primary name. This can be done just by looking at the synonyms and see whether a hit is found. The synonyms are mostly listed on the same website you found the gene in question. Else you can google the current name and see whether alternative hits are found and whether on that specific website the synonyms are mentioned.

In this case I was able to find a hit in Uniprot with "NADH pyrophosphatase" and since the gene name is mentioned that is associated with the enzyme in question I can use the gene name to annotate the enzyme in Wikipathways. With the gene name I was able to atomaticly annotate the enzyme. If the gene name was not found i can manually put the Uniprot ID with the specific enzyme/gene name.
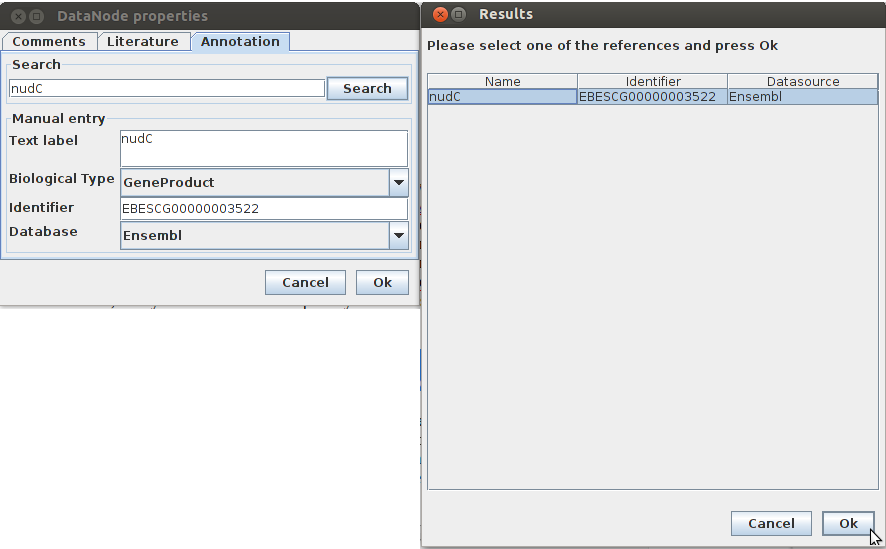
Now that I have annotated one of the data nodes that needed to be adjusted I can start a new one.
Abnormality in Wikipathways
There seems to be some kind of bug in Wikipathways. The bug is that (if we take the example above) that the ensembl identifier found for nudC does not exist when you go to the ensembl website. But since this is a bacteria if you go to "ensembl bacteria" a separate website of ensembl for bacteria, I was able to find the gene, but not the identifier, it seems that the identifier that is used in ensembl is completely different to the one annotated in Wikipathways:
Wikipathway ensembl identifier for nudC: EBESCG00000003522
Ensembl bacteria identifier:
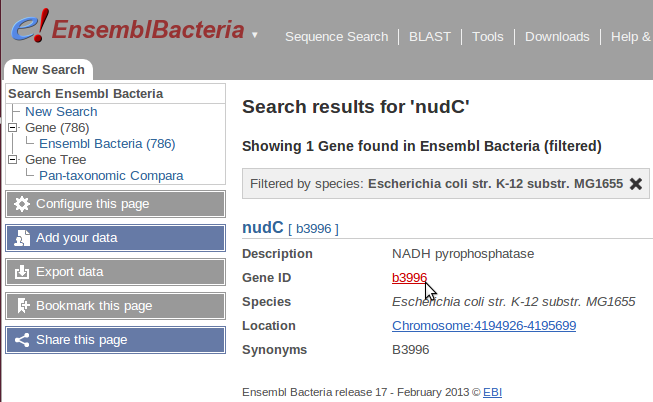
I reported this bug on Wikipathway-discuss, we will soon see what the cause of it is.
Interface RDF builder
I been working on the RDF building from commandline, but I seem to come to a hold because of a certain factor in the rdflib. I need to be able to change from "Literal" arguments to URIs easly without going into the code or writing a permutation of 3 on every combination possible. Currently i have a working code that does the job but its not command based yet. But I will be using:
import sys
sys.argv[argument]
This will allow me to take commands from a command base interface. But the previous explained problem can cause a problem if not defined correctly.
Fist we go to the the Wikipathway webpage: http://wikipathways.org/index.php/WikiPathways.
From there I entered the name of my pathway in the search engine.
After we entered the name and pushed "Search" we can add the species to narrow down the search.
Now I choose the to be adjusted NAD pathway (NAD salvage pathway I). After I have selected pathway you can edit them by pushing the "Edit pathway" button on the lower left of the pathway screen. Now I double click on the data node that I want to adjust.
So the first step is to try and find this enzyme/gene product a different database like ensembl (ensembl bacteria), uniprot etc. If like in this case we do not find a hit, we try to look whether it is a primary name. This can be done just by looking at the synonyms and see whether a hit is found. The synonyms are mostly listed on the same website you found the gene in question. Else you can google the current name and see whether alternative hits are found and whether on that specific website the synonyms are mentioned.
In this case I was able to find a hit in Uniprot with "NADH pyrophosphatase" and since the gene name is mentioned that is associated with the enzyme in question I can use the gene name to annotate the enzyme in Wikipathways. With the gene name I was able to atomaticly annotate the enzyme. If the gene name was not found i can manually put the Uniprot ID with the specific enzyme/gene name.
Now that I have annotated one of the data nodes that needed to be adjusted I can start a new one.
Abnormality in Wikipathways
There seems to be some kind of bug in Wikipathways. The bug is that (if we take the example above) that the ensembl identifier found for nudC does not exist when you go to the ensembl website. But since this is a bacteria if you go to "ensembl bacteria" a separate website of ensembl for bacteria, I was able to find the gene, but not the identifier, it seems that the identifier that is used in ensembl is completely different to the one annotated in Wikipathways:
Wikipathway ensembl identifier for nudC: EBESCG00000003522
Ensembl bacteria identifier:
I reported this bug on Wikipathway-discuss, we will soon see what the cause of it is.
Interface RDF builder
I been working on the RDF building from commandline, but I seem to come to a hold because of a certain factor in the rdflib. I need to be able to change from "Literal" arguments to URIs easly without going into the code or writing a permutation of 3 on every combination possible. Currently i have a working code that does the job but its not command based yet. But I will be using:
import sys
sys.argv[argument]
This will allow me to take commands from a command base interface. But the previous explained problem can cause a problem if not defined correctly.
The good thing about rdflib, it seems it does not write double triples. So I don't have to write a separate module to check for it.
Geen opmerkingen:
Een reactie posten